In terms of its structure, it is known that SARS-CoV-2 is a positive–sense and single-stranded ribonucleic acid (ssRNA) virus whose genome is approximately 30 kilobases (kB). Non-structural proteins play a crucial role in viral replication, whereby the four structural proteins of SARS-CoV-2 which include the spike (S), envelope (E), membrane (M) nucleocapsid (N) proteins, are encoded by the last third of the genome.
The N and S proteins of SARS-CoV-2 are the primary target for vaccine and therapeutic development through the production of monoclonal antibodies (mAbs) for neutralising infection. Several immune sera-derived polyclonal antibodies (pAbs) against SARS-CoV-2 are available, however there are few mAbs available for use to combat infection. It must be noted that mAbs are very specific and recognise a single epitope unlike pAbs which recognise multiple epitopes (READ MORE).
A recent paper by Mishra, et al., aimed to identify and describe the type of antibodies produced against nine SARS-CoV-2 proteins (Figure 1). In this present study, 18 fab-phage clones were identified and which bound to nine SARS-CoV-2 proteins.
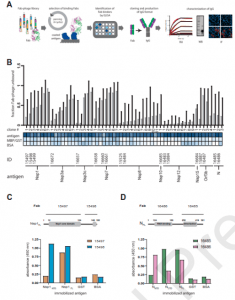
Figure 1: Synthetic antibody selection and characterization. (A) Schematic representation of the strategy in which antibodies were isolated by screening a phage-displayed Fab library for binding to SARS-CoV-2 proteins. Binding clones were identified by ELISA, transformed to IgG format, and characterized in vitro and in cells. (B) At the top, a plot of the % maximal Fab-phage binding signal in the presence of two different soluble antigen concentrations (gray: 250 nM, black: 50 nM) obtained by a two-point competitive ELISA. Fab-phage clones are ranked from lowest to highest signal ratios. Below the bar graph, Fab-phage clone numbers and binding signals to their cognate antigen obtained by ELISA is compared to negative controls (GST or MHT and BSA) and represented as a heat map with intensities ranging from high (blue) to low (white). At the bottom, the IDs for those antibodies converted to IgGs and their antigens are shown. (C) Fab 15497 and 15498 were measured for binding to immobilized Nsp1 N-terminal domain (Nsp1NTD), C-terminal domain (Nsp1CTD) and full-length protein (Nsp1FL). (D) Fab 15497 and 15498 were measured for binding to immobilized N N-terminal domain (NNTD), C-terminal domain (NCTD) and full-length protein (NFL). Negative controls (GST, BSA) were also included. Domain organization of Nsp1 and N are included and labeled above each panel in (A) and (B), respectively. Region of respective Fab binding are also indicated. (Mishra, et al., 2022).
In short the researchers suggest that mAbs availability may help improve the characterisation of host-viral interactions, in addition to improving our understanding of COVID-19 pathophysiology.
Journal article: Mishra, N., et al., 2022. Development of monoclonal antibodies to detect for SARS-CoV-2 proteins. Journal of Molecular Biology.
Summary by Stefan Botha










