Tuberculosis (TB), caused by members of the Mycobacterium tuberculosis (Mtb), is a severe chronic disease that can affect humans and animals. Mtb is a leading cause of death in the world (1.5 million deaths every year). Among people who are infected, only 10% will develop TB, 90% of these infections are latent (LTBI). This pathology presents the difference in immune control of Mtb. Macrophages (MΦs) are significant phagocytes that play a key role in innate and adaptive immunity. Their phenotypes stimulated with IFNy (pro-inflammatory) or IL-4, IL-13, and IL-10 (anti-inflammatory) determine the type of Mtb response.
In a recent paper, Khan, et al., hypothesized that the MΦ phenotype can enhance anti-TB function following cytokine stimulation, and this stimulation can explain the development of LTBI (Figure 1). They showed that infected M1-MФs survived better with upregulation of genes and pathways that regulate antigen processing Compared to M2-MФs. They highlight also M1-MΦs driven with IFN-γ increased the ability to bacterial control with the pro-inflammatory cytokine, nitric oxide expression, and autophagy-dependent degradation of Mtb, whereas IL-4 programmed M2-polarized MΦs (M2-MΦs) are permissive for Mtb proliferation.
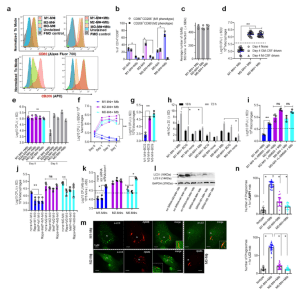
Figure 1: Human umbilical cord and peripheral blood-derived macrophages show heterogeneity in mycobacterial killing associated with oxidants and autophagy. Human cord blood (CBM) or healthy donor PBMC-derived MФs were cultured in the presence of either recombinant human IFN-γ (M1; 10 ng/mL) or human IL-4 (M2; 10 ng/mL) for 5 days and rested for 2 days. Untreated cells were M0-MФs. a, b Surface expression of receptors by CBM- derived naïve and Mycobacterium tuberculosis (Mtb; H37Rv)-infected M1- (CD80+/CD206−) and M2-MФs (CD80−/CD206+) on day 3 using flow cytometry and quantitation (*p < 0.01 t test); gating strategy shown Supplemental Fig. 1. c CBM-derived differentiated MФs were infected with Mtb for 4 h followed by microscopic counts of rfp-labeled MtbH37Rv to determine uptake verified using CFU counts. d PBMC-derived MФs from five healthy donors were differentiated using the indicated cytokines followed by infection with Mtb and CFU assay on day 4. Each point represents one donor (**p < 0.05; Kruskal–Wallis test). e PBMC-derived MФs were differentiated using GM-CSF (M1), IL-4 (M2a), IL-1β M2b), IL-10 (M2c) or left intreated (M0) followed by Mtb infection and CFU assay on day 4 (**p < 0.007). f CBM-derived, cytokine differentiated MФs were infected with Mtb followed by CFU assay over time (**p < 0.006; data from 3 experiments shown). g CBM-derived, differentiated MФs were infected with M. bovis BCG followed by CFU assay on day 4 (**p < 0.005). h CBM-derived and differentiated uninfected MФs or those infected with Mtb or BCG were incubated and at indicated time points, cultures were tested for nitrite using diaminofluorescein diacetate and fluorometry (*,**p < 0.005, t test). QPCR for mRNA of iNOS and protein are shown in Supplemental Fig. 3a and reactive oxygen species levels in Supplementary Fig. 3b. i MФs infected with Mtb as in panel h were incubated in NMMA (0.5 mM; N-monomethyl L-arginine) followed by CFU assay on day 3 (**p < 0.009). j CBM-derived, differentiated MФs infected with Mtb were incubated with 10 μM Rapamycin, 100 μM Metformin or their combination followed by CFU assay on day 3 (**p < 0.009). k CBM-derived, M1-, M2- and M0-MΦs were treated in the presence or absence of siRNA vs. beclin1 (ATG6) or its scrambled control followed by infection with Mtb and CFU counts on day 3 (*p < 0.007). Blot validation of Knockout using siRNA vs. beclin1 (ATG6) is shown in Fig. 3h. l MΦ lysates of panel k collected at 18 h were analyzed using western blots for the lipidation of microtubule-associated light chain 3 (LC3). Lipidation is indicated by LC3-II. m CBM-derived MΦs were infected with rfpMtbH37Rv and stained for an LC3 autophagy marker or LAMP1 lysosome marker using specific antibodies, Alex-Fluor485 conjugates, and imaged using confocal microscopy. Panels illustrate LC3 colocalization; LAMP1 stains using gfpMtbH37Rv is shown in Supplementary Fig. 3c. n Quantification of phagosomes colocalizing with LC3 are shown using an N90 Nikon fluorescence microscope (IF) and Metaview software (*p < 0.004, t test). For panels (b–c–e–f–g–h–i–b–k–n), p-values were calculated using a one-way ANOVA with Tukey’s post-hoc test; one of 2–3 similar experiments shown. CFU or IL-2 assays had triplicate wells plated per group or donor. Panels (d, g, i–k) horizontal dotted lines indicate day 0 CFU (4 h post-infection CFU). All Mtb CFU experiments used MOI of 1.
They found that positive autophagy regulation in Mtb-infected M1-MΦs strongly expressed Sirtuin5, HDAC2 (positive autophagy regulator). However, the expression of Sirtuin2 (negative autophagic regulator) Mtb-infected M2-MΦs was correlated to bacterial survival. They finally suggested that M1 and M2 phenotypes can mediate anti-TB response in humans and macaques. The gene expression difference could be used as Host directed therapeutic targeting MΦs polarization.
Journal article: Khan, A. et al., 2022. Human M1 macrophages express unique innate immune response genes after mycobacterial infection to defend against tuberculosis. Communications biology.
Summary by Mberkadji Ngana










