In recent times, the gut microbiome has become the focal point of many research groups. There intestinal microbiome consists of a plethora of bacterial species, all existing in an intricate balance whereby this balance is key to overall good health for the individual. These bacteria make up the aptly named intestinal flora, integral for digestion and for keeping infection at bay. These bacteria are non-pathogenic and are tolerated by the immune system. The mechanism used by the immune system to maintain the delicate balance in the intestine remains unknown. It has been reported that type A immunoglobulins i.e. IgA antibodies, play an important role in the maintenance of this balance. These molecules for a natural part of the immune system, and by means of a lock-and-key principle, identify specific exogenous pathogens. Dimeric IgA is secreted across the mucous membranes in response to non-pathogenic taxa of bacterium which accounts for most antibody production in mammals. An imbalance of microorganisms may have a detrimental effect on health and cause intestinal diseases.
Researchers from the Department of BioMedical Research (DBMR) at the University of Bern and collaborators have recently shown in a murine model that IgA antibodies regulate and modulate, at several levels, the activity of non-pathogenic bacteria. This allows for the regulation of the immune system, aiding in the maintenance of the balance of microbes within the gut microbiome. Rollenske, et al., demonstrated that the immune system recognises and restricts specific bacteria in a highly specific manner (For more see – Regulation of Immunity & the Microbiome).
The way in which IgA antibodies regulate the coexistence of microbes in the intestine has remained unsolved. Possible reasoning would be that the study of IgA antibodies under physiological conditions in vivo was not possible. In this present study, the researchers were able to develop a method to overcome this challenge. They produced IgA antibodies specifically directed against a type of Escherichia coli, a typical intestinal bacterium. The antibodies were able to recognise and bind on the membrane of the microorganism.
Following three years of research and work, Rollenske, et al., were able to accurately track the in vitro and in vivo effects of IgA antibodies in the intestines of germ-free mice. Through the use ofrecombinant dimeric monoclonal IgAs (mIgAs), the authors mapped the intestinal plasma cell response to microbial colonization with a single microorganism in mice, identifying a range of antigen-specific mIgA molecules targeting defined surface and non-surface membrane antigens.
It was reported that IgA antibodies affect bacteria in several ways including: restricting the mobility of bacteria, microbial carbon-source uptake, bacteriophage susceptibility, membrane integrity and regulating the metabolism of the bacteria (Figure 1). Even in cases in which the same microbial antigen is targeted, microbial metabolic alterations differed depending on IgA epitope specificity. The response elicited is dependent on the antigen presented on the bacterial surface being specifically recognised by the immune system. This meaning that the immune system is able to influence the benign intestinal bacteria through different approaches on a simultaneous basis. This leading to a concept of IgA parallelism.
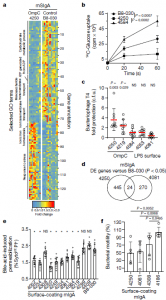
Figure 3: Surface-binding mSIgA exerts antigen-dependent individual and generic functional alterations in intestinal bacteria. a, Differential gene expression of enriched selected GO term gene members in faecal JM83 bacteria following reconstitution with OmpC-binding mSIgA4250 as compared with negative control mSIgAB8-030. Numbers (right y axis) identify gene annotations in Supplementary Table 7. b, [14C]Glucose uptake in ΔompF bacteria coated with indicated mIgAs, (x ± s.d., mSIgAB8-030 n = 6, mSIgA4250 n = 5, mSIgA4584 n = 3). cpm, counts per minute. c, Protection from bacteriophage T4 infection of mIgA-coated JM83 bacteria according to the category of the mIgA target (x ± s.d., n = 9 for each condition). d, Venn diagram of differentially expressed (DE) genes at 10.5 h after mSIgA4250 and mSIgA4081 reconstitution, both compared to mSIgAB8-030. e, Flow cytometric analysis of faecal JM83 bacteria after exposure to bile acids. Percentages (y axis) of dying bacteria are shown (gating in Extended Data Fig. 9a) after pre-incubation with mIgA (x ± s.d., n = 12 for each condition). Source data show exact P values. f, JM83 bacterial plate motility assay after pre-incubation with indicated mIgA (x ± s.d., n = 5 for each condition). Statistics show two-sided unpaired Student’s t-test (b, c, e, f), *P < 0.05; NS, not significant. Data are pooled from two (b, c, e, f) or five (a, d) independent experiments (Rollenske, et al., 2021).
“…clonal selection into the plasma cell IgA repertoire induced by a single microbe provides a series of SIgA-mediated parallel mechanisms, including metabolic modulation, protection from bile acids or bacteriophage, and motility alterations; these allow the immune system to exert controls in the intestinal lumen that fine-tune host–microbial mutualism. Parallelism occurs in two ways: first, the response to a single microbe has different components on distinct targets with a series of different functional results; and second, one mSIgA can have more than one functional result… In these studies, we measured a series of functional SIgA responses on a single microbe that was also used as the inducing agent…”
The findings from this study build on our basic understanding of the immune system in the intestine. In addition, it has provided insight into how this understanding may contribute to the development of vaccines.
Read more at Immunopaedia – A Snapshot of the Immune System
Journal Article: Rollenske, et al. 2021. Parallelism of intestinal secretory IgA shapes functional microbial fitness. Nature.
Summary by Stefan Botha










