In a recent study, researchers have demonstrated that mucosal-associated invariant T (MAIT) cells, a form of immune cells, play several complex roles during diseased and healthy conditions (Figure 1). The results provide a foundation on which to develop research on MAIT cells, using them as targets for immunotherapies and vaccines.
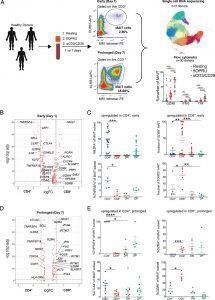
Figure 1: MAIT cell CD4 or CD8 expression levels are associated with distinct transcriptional signatures. (A) Experimental schematic of the MAIT cell in vitro activation assay used for single-cell RNA sequencing (scRNA-seq) and flow cytometric analyses. A total of 30 healthy donor PBMCs were assayed by flow cytometry after 15 h (day 1; early activation) or 7 d (day 7; prolonged activation) of incubation under resting, 5-OP-RU, or anti-CD3/CD28 conditions. A subset of three of these donors underwent FACS for scRNA-seq under the same conditions. All conditions included IL-2 and a subset of day 7 conditions also included TGF-β. (B) Differentially expressed genes in CD8+ [+ log fold change (FC)] or CD4+ MAIT cells (− log FC) after 15 h of incubation in the resting condition. The y-axis shows log FDR-adjusted p value with a significance level of FDR-adjusted p < 0.05. (C) Mean % ± SEM of CD25, OX40, GZMB, and EOMES staining in CD4+, CD8+, DN, or DP MAIT cell subsets measured by flow cytometry in the same condition as (B). Represents three independent experiments. n = 5–30 donors. (D) Differentially expressed genes in CD8+ or CD4+ MAIT cells after 7 d of incubation in the resting condition. Same axes as panel B. (E) Mean % ± SEM of OX40, CTLA4, GZMA, and GZMK staining in MAIT cell subsets measured by flow cytometry in the same condition as (D). Represents two independent experiments. n = 5–10 donors. Flow cytometric statistical comparisons were made by an unpaired t test, and reported p values are adjusted for multiple comparisons using the Holm–Sidak method. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001. DN, double-negative (CD4−CD8−); DP, double-positive (CD4+CD8+); GZMA, granzyme A; GZMB, granzyme B; GZMK, granzyme K (Vorkas, et al., 2022).
MAIT cells are extremely abundant in the human body. They can be activated by non-peptide vitamin intermediates from microbes and are involved in both infectious and non-infectious diseases mechanisms and process.
In this present study they made use of single cell RNA sequencing technology as well as other immunologic techniques to identify one cell type with a semi-invariant T cell receptor, further demonstrating MAIT cell heterogeneity that places emphasis on conventional T cell biology. They also demonstrated this heterogeneity in response to infection with M. tuberculosis in humans (Figure 2).
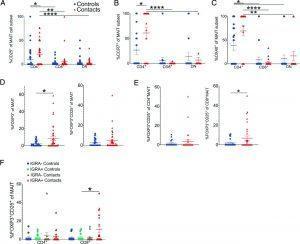
Figure 2: MAIT cell subsets differentially respond to M. tuberculosis. (A–C) Mean % ± SEM of CD25 (A), CCR7 (B), and OX40 (C) staining of CD4+, CD8+, or DN MAIT cell subsets in healthy household contacts of TB patients (red triangles) compared with healthy unexposed community controls (blue circles). The same legend also applies to (D) and (E). Results represent more than three independent experiments. n = 18–41 donors. (D) Mean % ± SEM of FOXP3+ (left) or FOXP3+CD25+ (right) MAIT cells. Results represent more than three independent experiments. n = 36–42 donors. (E) Mean % ± SEM of FOXP3+CD25+ CD4+ (left) or CD8+ MAIT cells (right) in the same donors as (D). (F) Mean % ± SEM of FOXP3+CD25+ CD4+ or C8+ MAIT cells stratified by IGRA status in the same donors as (D). Flow cytometric statistical comparisons were made by Mann–Whitney U test in (A)–(C) or unpaired t test panels (D)–(F), and reported p values are adjusted for multiple comparisons using the Holm–Sidak method. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001. IGRA, M. tuberculosis–specific IFN-γ release assay (Vorkas, et al., 2022).
The researchers reported that the heterogeneity of MAIT cells includes distinct CD4+ and CD8+ populations, in addition to killer, helper, and regulatory cell phenotypes. This highlighting the complexity of MAIT cells and their function.
In their own words:
“In summary, we present an integrated transcriptional and immune landscape of human MAIT cell activation at single-cell resolution. Our results provide a detailed analysis of novel MAIT cell clusters defined by early and prolonged stimulation, including distinct CD4+ and CD8+ MAIT cell signatures. We also show that these phenotypes are recapitulated in the setting of early M. tuberculosis exposure and infection. This functional map of MAIT cell activation biology will help inform future investigations into how MAIT cells specialize in healthy and disease states.”
Journal article: Vorkas, C.K., et al. 2022. Single-Cell Transcriptional Profiling Reveals Signatures of Helper, Effector, and Regulatory MAIT Cells during Homeostasis and Activation. Journal of Immunology.
Summary by Stefan Botha










