In this webinar researchers from the University of Ghana: Joyce Ngoi and Peter Quashie discuss the analysis of SARS-CoV-2 sequences which include imported and circulating viruses at the inception of Ghana’s outbreak, and circulating viruses 2-3 months after Ghana’s first reported cases. (Source: IUIS Webpage)
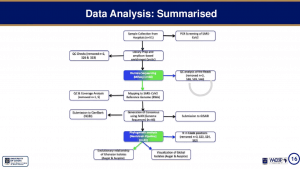
Joyce Ngoi’s talk began with a brief introduction on SARS-CoV-2 epidemiology which showed that after the first reported case SARS-CoV-2 infection on 12th March community transmission has resulted in >45000 cases by 24th August (>46000 cases on 16th Sep 2020). She then explained that utility of genomic epidemiology* for track community transmission and evolution of emerging viruses. This was followed by an overview of the workflow and analysis pipeline used to generate results:
- Sample Processing [RNA extraction and amplification]
- Sample selection of samples with amplified RNA
- CDNA preparation & Virus enrichment to further amplify genetic material required for sequencing
- Library construction, barcoding of sample enabling sequencing of pooled samples
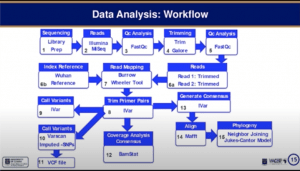
Peter Quashie then took over the presentation discussing the analysis workflow required to generate results (see below).
Results demonstrated that early stages of the SARS-CoV-2 transmission were predominantly from strains imported from “Europe and US”. Phylogenetic analysis suggested lower viral diversity (clades) in Ghana compared to the rest of Africa. He also showed that as seen in other regions viral strain D614G quickly became one of the dominant strains. He also presented data that described novel mutation suggesting instances of local evolution of new viral genotypes virus. One example was the identification of genotype L84S, also detected in India, that also possessed additional mutation that seems to be specific to strains circulating in Ghana.