November 18th 2020, marked the end of the most recent Ebola virus disease outbreak in the Democratic Republic of Congo. Though this outbreak is magnitudes smaller than the most severe Ebola outbreak (predominantly in Guinea, Liberia and Sierra Leone) which lasted almost three years (2013-2016) and caused more than 28000+ cases, the fatality rate was still high >40%. Despite the availability of vaccines and antibody-based immunotherapy, the case fatality rate of treated patients is still very high. Thus, highlighting the need for an improved understanding of Ebola pathogenesis to improve the current interventions we have. Unfortunately, Ebola animal models are challenging to perform. Firstly, the progression of Ebola in mice, the most commonly used animal model for understanding disease pathology, does not entirely resemble human pathogenesis. Secondly, though non-human primates are a better animal model, Ebola studies are often based on a small sample size and require large biosafety level 4 facilities due to the lethality of Ebola. Further differences in anti-Ebola immunity observed between in vivo and in vitro studies possess additional challenges.
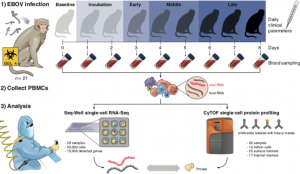
Figure Legend: Under BSL-4 containment, we collected blood samples from a total of 21 rhesus monkeys at multiple days post-EBOV inoculation, extracted peripheral blood mononuclear cells (PBMCs), and profiled single-cell transcriptomes and 42 protein markers using Seq-Well and CyTOF. Seq-Well quantifies both host (black) and viral (red) RNA expression, allowing comparisons between infected and bystander cells. Daily clinical parameters (body temperature, clinical signs, and body weight) were also collected for each animal, and complete blood counts were obtained for each blood draw. (Source: Kotliar et al., 2020.)
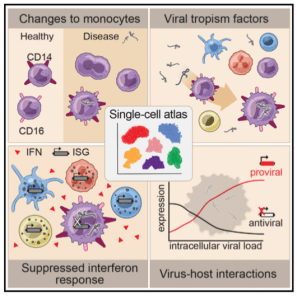
In a recent Cell article, Kotliar et al., utilised RNA-sequencing (RNA-seq) and mass cytometry to immune profile 90 peripheral blood mononuclear cell (PBMC) samples from 21 rhesus monkeys before or during lethal Ebola virus challenge in vivo. This study represents the first BSL-4 study to investigate pathogenic immunity using high dimensional single-cell technologies. They demonstrated that of blood cells phenotype, monocytes undergo the most significant gene expression changes during Ebola. One of the most striking observations was a decrease in genes associated with antigen presentation, which was not only restricted to monocytes infected with Ebola virus-infected but to by-stander monocytes as well. Reduction of such genes could explain why “failed or delayed adaptive immunity is a hallmark of fatal Ebola disease in humans”. They also showed that Ebola infection is associated with increase in highly proliferative “immature” CD14-CD16- (double negative) monocytes with transcriptomes more similar to bone marrow monocyte precursors than circulating monocytes in both NHPs and humans. Lastly, they showed that viral infection was associated with selective down-regulation of anti-viral (IFN-stimulating genes) and up-regulation of pro-viral genes in Ebola virus infected monocytes. The authors also investigated other aspects of Ebola immunity which are not covered in this summary.
Though study did have some limitations, such as not investigating tissue-specific pathology and predominantly focusing on monocyte immunology. The study does highlight the utility of single-cell immuno-phenotyping in understanding Ebola pathogenesis and immunity, even for BSL-4 experiments.
Journal Article: Kotliar et al., 2020. Single-Cell Profiling of Ebola Virus Disease In Vivo Reveals Viral and Host Dynamics. Cell
Summary by Cheleka AM Mpande