Researchers from Sweden have described the variant formation of Trypanosoma cruzi which are capable of evading the immune system and causing disease. The results from this study provide new insights into the treatment, prevention and diagnosis of the Chagas disease, which is one of the neglected and highest impact parasitic infections in the Americas. This disease is caused by Trypanosoma cruzi infection, causing damage to the gastrointestinal tract and heart. Due to genetic variations, the parasite is able to evade the immune system and cause disease.
The researchers in this study were able to identify how Trypanosoma cruzi can create genetic variation through hybridisation between different strains. Through genome mapping of parental lineages and offspring using high throughput and large-scale DNA sequencing techniques, they were able to develop a picture to describe these variations (Figure 1). They also further described genetic recombination events that occur which lead to these hybrid strains.
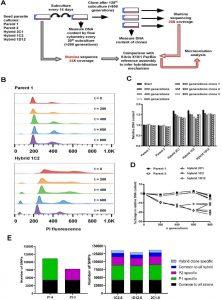
Figure 1: In vitro microevolution of Trypanosoma cruzi hybrids. (A) Overview of experimental design. (B) DNA content profiles shown as propidium iodide (PI) staining intensity histograms measured by flow cytometry. PI fluorescence = relative fluorescence units; y-axis shows number of events from the population of parasites at the indicated generation of the in vitro culture experiment from start (t=0) to finish (t=800). Plots of one parent and one hybrid are shown, representative of three biological replicates for each time point. The vertical dashed line marks the position of the G1 fluorescence intensity peak at t=0. (C) Quantitative analysis of DNA content for both parents and three hybrid strains at each time point and for three clones per strain generated after the final time point of 800 generations. Data are the mean + SEM of three biological replicates after normalization against an internal standard and conversion to a ratio of the mean of the two parents at t=0. The predicted total DNA content ranges equating to tetraploid (4n1k) and triploid (3n1k) parasites are indicated, under the assumption that kDNA constitutes between 20% and 30% of total DNA. (D) The same data as shown in C, represented as the % change in DNA content over time compared to t=0. (E) Distinct genomic signatures exclusive to each parental strain were inherited by the hybrid strains. In black are represented SNPs present in both parental strains, in green SNPs exclusive to the P1 strain, in purple SNPs exclusive to the P2 strain, in dark blue SNPs common to all hybrids, and in light blue SNPs exclusive to each hybrid clone (Matos, et al., 2022).
Journal article: Matos, G.M., et al., 2022. Experimental microevolution of Trypanosoma cruzi reveals hybridization and clonal mechanisms driving rapid diversification of genome sequence and structure. eLife.
Summary by Stefan Botha










