It has become a global effort for researchers to try and identify the underlying mechanisms of transmission, the ability to evade immunity, and many other aspects of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).
Severe SARS-CoV-2 infection is prevented, in part, by the function and activity of T cells. The ability of the SARS-CoV-2 variants, in addition to the variants of concern (VOCs), to evade the immune response due to mutations can be halted by either vaccination or natural infection, whereby most T-cell epitopes are conserved. T-cell responses remain unaffected by mutations, providing some protection for the host against VOCs and wildtype SARS-CoV-2. Therefore, it is important for us to understand the factors that contribute to frequency and activity of the T-cell response during SARS-CoV-2 infection.
Previous studies have shown that activation of CD4+ and CD8+ T-cells is inversely correlated with COVID-19 severity and death, highlighting a strong Th1-bias which is beneficial for recovery form COVID-19.
In a new study by Binayke, et al., the researchers investigated the innate and adaptive immune responses during both the late and early stage of SARS-CoV-2 infection. Additionally, they wanted to establish the relationship between early and late stage infection and the immune responses during mild infection with SARS-CoV-2. Performing a longitudinal metabolic analysis of SARS-CoV-2-infected patients they revealed distinct metabolites that show correlation with the innate and adaptive immune responses (Figure 1).
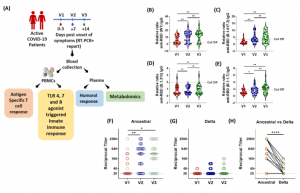
Figure 1: Longitudinal Analysis of Humoral Immune Response against the SARS-CoV-2 during acute COVID-19 infection. (A) Schematic experimental design and longitudinal sample collection at specific time intervals after confirmed SARS-CoV-2 infection in mildly infected patients. (B-E) The longitudinal anti-RBD IgG responses were evaluated by performing ELISA against the RBD proteins of (B) WT (Wuhan isolate) (C) Delta (B.1.617.2) (D) Beta (B.1.315) (E) Alpha (B.1.1.7). All data, represented as ratio-converted ELISA reads to a pool of pre-pandemic negative control samples (relative ratio), were plotted using violin plots. (F-H) The longitudinal neutralizing antibody titers against (F) the ancestral strain and (G) the delta strain of SARS-CoV-2 during V1 (day 0-3), V2 (day 7) and V3 (day 14) from COVID19 positivity (H) The paired representation of NAb titers in the active COVID-19 patients during V3 against the ancestral and delta variants of SARS-CoV-2. (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission (Binayke, et al., 2022).
The study was revealed several limitations, including a small sample size as well as the inability to determine the levels of type 1 IFN following TLR stimulation of PBMCs.
This study showed that PBMC-derived innate immune responses are key for activating a strong T-cell response to infection, however, the length of this response lasts.
NB to note: bioRxiv is a preprint server which publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, or guide clinical practice or treated as established information.
Journal article: Binayke, A., et al. 2022. Innate immune response and distinct metabolomic signatures together drive and shape the SARS-CoV-2-specific T cell response during COVID-19. bioRxiv.
Summary by Stefan Botha










