In a recent paper, researchers aimed to characterise the T-cell antigen response in response to SARS-CoV-2 variants using the ATLAS screening assay (READ MORE). Mutation of the receptor-binding domain (RBD) and the receptor-binding motif (RBM) has led to immune response escape and vaccine induced immunity escape by SARS-CoV-2. The need understand the mechanisms underlying this escape, will aid researchers in the development of new therapeutics combatting the infection.
In this present study, they collect the PBMCs (Peripheral blood mononuclear cells) from various cohorts and unexposed controls. The ATLAS assay was utilised to quantify CD4+ and CD8+ T-cells through analysing the pathways of HLA class I and II. Using an ATLAS library (Figure 1), they were able to profile each of the cohorts.
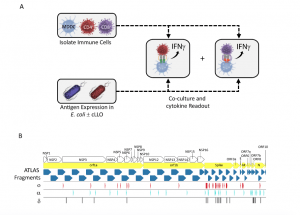
Figure 1. The ATLAS assay and library design. (A) Each putative antigen is expressed individually in two E. coli strains one with and one without cLLO (bottom left box), which are arrayed in expression libraries. Monocyte derived dendritic cells (MDDCs) as well as CD4 + and CD8 + T cells are isolated from peripheral blood and co-cultured overnight with the E. coli libraries and IFNγ output measured in each well. (B) Schematic representation of the library design depicting the location of the ATLAS Fragments (blue chevrons) in relation to the SARS-CoV-2 orfs (yellow chevrons) and orf1ab NSPs (white chevrons). Mutations from the Omicron (ο), Delta (δ), and Alpha (α) VOCs are shown as red, cyan, and black barcodes, respectively (Foti, et al., 2022).
The researchers were able to describe and identify T-cell responses throughout the SARS-CoV-2 proteome, subsequently identifying and characterising several conserved/common regions of antigens in agreement with literature. They further showed that the ATLAS assay further optimises the use of the immunoproteasome for peptide analysis.
In their own words:
“ATLAS revealed several common or frequent antigenic regions as well as an abundance of responses in the unexposed cohort potentially the result of pre-exposure to related coronaviruses. ORF10 was a common CD4 + response in the unexposed cohort while spike was identified as a common and frequent target in both cohorts. Moreover, the spike response profiles allowed us to accurately predict the impact of Omicron spike mutations. This analysis could thus be applied to study the impact of future emerging VOCs.”
In short, understanding the effects of Omicron mutations as well as other variants and how they affect the immune response, will aid us in our fight against the SARS-CoV-2 virus.
NB to note: bioRxiv is a preprint server which publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, or guide clinical practice or treated as established information.
Journal article: Foti, J. J., et al., 2022. The ATLAS™ screening assay reveals distinct CD4+ and CD8+ SARS-CoV-2 antigen response profiles which have implications to Omicron cellular immunity. bioRxiv.
Summary by Stefan Botha










