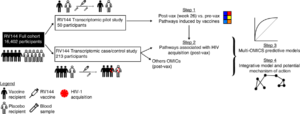
Schematic of Fourati et al., study overview. Briefly, Four analysis steps were used to identify transcriptomic markers of risk of HIV-1 acquisition among RV144 vaccinees. A first transcriptomic dataset of blood collected from 40 HIV-1 negative vaccinees and 10 HIV-1 negative placebo recipients prevaccination and 2 weeks after vaccination was used to identify pathways modulated by the RV144 vaccine (step 1). A second independent transcriptomic dataset of blood collected from 183 case–control vaccinees (including 31 infected participants) and 30 placebos (including 17 infected participants), 2 weeks after vaccination was used to identify pathways associated with HIV-1 acquisition. Logistic regression was used to build a multi-OMICS classifier of HIV-1 acquisition among RV144 vaccinees (step 3) and a projection-based integrative analysis was used to associate the different OMICS to identify mechanistic mediators of vaccine response (step 4). Source: Fourati et al., Nat. Comms 2019
The RV144 trial, infamously known as the “Thai trial”, is the only HIV vaccine to show 31% efficacy against HIV acquisition. Based on results of immune-correlates analysis, Haynes et al., hypothesised that vaccine induced IgA antibodies (Abs) outcompete vaccine IgG responses resulting in increased HIV acquisition. Additional analysis by Lin et al., showed that polyfunctional HIV-specific CD4+ T cells were associated with reduced risk of HIV acquisition. A study recently published by Fourati et al., further contributes to this body of research, identifying vaccine induced transcriptomic pathways that are associated with increased risk of HIV acquisition.
Gene set enrichment analysis of HIV-envelope stimulated PBMCs revealed that the RV144 vaccine induced genes associated with antigen presentation and antiviral functions. Specifically, Fourati et al., linked IFN-γ responses with reduced risk of HIV acquisition, while NFKB and mTOR signalling pathways were associated with increased risk of acquisition. Signalling by NFKb and mTORC1 lead to activation genes required for HIV replication as well as proliferation of HIV-1 target cells, mechanistically highlighting how this could result in increased HIV acquisition.
Fourati et al., further integrated results from gene expression, antibody and PBMC-ICS analysis. They showed that IFN-γ responses identified by transcriptomics correlated with env-specific IgG and CD4 T cells responses. Researchers hypothesise that the RV144 vaccine vector (ALVAC) triggered plasmacytoid dendritic cells to induce type I interferons, which intern induced expression of the Fc gamma receptor facilitating IgG mediated antibody dependent cellular mediated cytotoxicity. Additionally, they showed that enrichment of genes associated with antigen presentation correlated with activated CD4 T cells, which could potentially provide help to B cells to produce IgG antibodies.
In summary, Fourati et al., used an unbiased systems biology approach to identify transcriptomic signatures associated with risk of HIV acquisition. These findings highlight the importance of post-hoc analyses in revealing immune pathways that can be targeted by future HIV vaccines.
Journal Article: Fourati et al., 2019. Integrated systems approach defines the antiviral pathways conferring protection by the RV144 HIV vaccine. Nat. Communications











