In a new study, researchers have successfully created the first comprehensive map (Figure 1) of the human immune system, illustrating all connections in full and describing the communication between immune cells and how we can use this as a tool to investigate disease and develop new therapeutics. showing how immune cells communicate with each other and ways to modulate these pathways in disease.
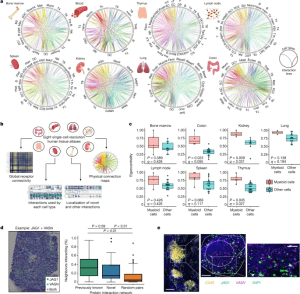
Figure 1: An interactive atlas of immune cell connections across the human body. a, Systematic integration of single-cell datasets to map cellular connectivity across tissues with substantial immune populations. Cell types are positioned around each circle, with each position along it marking a cell-surface protein. Linkages formed by physically interacting surface proteins between cell types are marked by curved lines, coloured by interaction identity. Full abbreviations are listed in Supplementary Table 5. b, Functionalities available through our interactive atlas of physical immune interactions (https://www.sanger.ac.uk/tool/immune-interaction/immune-interaction). c, Myeloid cells act as interaction hubs in immune tissues. Eigenvector centrality metrics of myeloid cells compared to all other populations after converting the total interaction count for all cell–cell pairs into a weighted undirected graph. Data are shown as Tukey box plots with Benjamini–Hochberg P values calculated from a two-sided Welch’s t-test. d, Spatial transcriptomics of a human lymph node confirms that our identified interaction partners are physically colocalized in situ. An example data point of a tissue section analysed for the JAG1 + VASN interaction is shown. The percentage of measured spots in which the expression of one protein of an interacting pair is spatially connected to the other protein of the pair is compared for previously reported interactions (green), novel interactions (blue) and a negative control of the same proteins with interaction links randomly permuted (yellow) (n = 100). Data are shown as Tukey box plots with P values calculated from a Tukey’s honest significance test. e, Single-molecule RNA hybridization on human lymph nodes defines regions in which newly identified interaction partners are expressed in spatially bordering cells. A single lymphoid follicle enriched in CD45+ leukocytes is magnified (left), showing the zonation of JAG1- and VASN-expressing cells into the corona and the germinal centre, respectively (middle). An inset (right) highlights a region of bordering cells expressing each marker. Scale bars, 200 μm (left); 100 μm (middle); 50 μm (right).
The immune system comprises several different types of cell which each have a unique function. Some of these cells localise to the site of injury or infection and once detected, essential messages are communicated to other cells of the body in order to induce an efficient immune response. This cell-to-cell signalling can be facilitated through surface receptors which are proteins on the surfaces of cells that bind to other proteins. Understanding how cells communicate with each other and when, is essential for developing new treatments and improving existing ones such as immunotherapies, to enhance the immune response against disease.
Importantly, this study also provides insight into the mechanisms that may underlie autoimmune conditions and how we may go about treating them in a more efficient way.
In short, this study has provided a unique and powerful tools for researchers and clinicians to use to fight disease, autoimmune disorders, develop novel drugs for disease treatments and improve immunotherapies for patients with diseases such as cancer.
In their own words:
“To our knowledge, our study is among the first to systematically map and model how the collective actions of individual receptor molecules through physical laws could explain and predict cellular connectivity on a scale as large as the circulating immune system. Our analysis and the methods that we developed provide a template for future studies looking at physical cell wiring networks in detail. From these combined approaches, we may finally begin to disentangle cellular circuits in immunity and beyond, bridging from individual protein molecules to multicellular behaviour.”
Journal article: J. Shilts, Y. Severin, F. Galaway, et al. A physical wiring diagram for the human immune system. Nature.
Summary by Stefan Botha










