Mode of Action
- Mode of Action - NRTI
- Mode of Action - NNRTI
- Mode of Action - Protease Inhibitors
- Mode of Action - Integrase inhibitor
- Mode of Action - Entry Inhibitors
Mode of Action – NRTI
Nucleoside Reverse Transcriptase Inhibitors (NRTIs) inhibit reverse transcription by causing chain termination after they have been incorporated into viral DNA. For these drugs to be active they need to be phosphorylated intracellularly. This was the first group of antiretroviral agents to be used against HIV.
Usually two drugs from this class are used to form the backbone of HAART.
Mechanism of Action
Mechanisms of HIV antiretroviral drug action and drug resistance
Nucleoside reverse transcriptase inhibitors (NRTI)
Mechanism of action
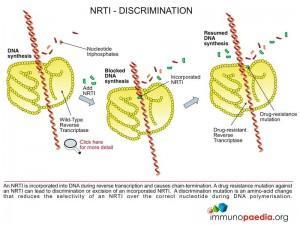
The NRTI class of antiretroviral drugs are chemical compounds that are nucleotide base analogues. They function as chain-terminators during the extension of the DNA chain during the reverse transcription process which is carried out by HIV reverse transcriptase. The NRTI compounds permit correct base-pairing and incorporation into the DNA chain, however, an important hydroxyl group required for addition of the next nucleotide has been replaced by a non-reactive chemical group.
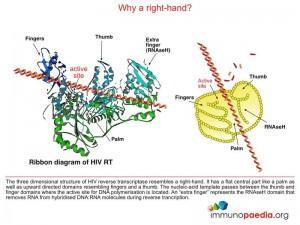 With the exception of Tenofovir, the NRTI compounds that are taken up by the cell do not contain phosphate groups and require three phosphate groups to generate the triphosphate form of the base analogue before it can be used as a substrate by reverse transcriptase. Tenofovir contains a single phosphonate group to which only two phosphate groups need to be added to generate the active compound. The additional phosphorylation is carried out by host cellular enzymes (kinases).
With the exception of Tenofovir, the NRTI compounds that are taken up by the cell do not contain phosphate groups and require three phosphate groups to generate the triphosphate form of the base analogue before it can be used as a substrate by reverse transcriptase. Tenofovir contains a single phosphonate group to which only two phosphate groups need to be added to generate the active compound. The additional phosphorylation is carried out by host cellular enzymes (kinases).
Mechanism of Resistance
Mechanisms of HIV antiretroviral drug action and drug resistance
Nucleoside reverse transcriptase inhibitors (NRTI)
Mechanism of resistance
There are two ways in which NRTI resistance can be achieved:
- Discrimination pathway
- Excision pathway
1. Discrimination pathway
These resistance mutations are amino acid changes in the primary structure of reverse transcriptase that increase the selective ability of the enzyme to incorporate the natural nucleotide over the NRTI-triphosphate (eg. K65R, K70E, L74V, M184V and Q151M).
There are two ways in which discrimination can be achieved:
- Decrease the binding affinity of the NRTI-triphosphate over the natural nucleotide in the reverse transcriptase active site (eg. M184V and V75T). In experiments with wild-type and mutant reverse transcriptase carrying the M184V mutation measuring the binding affinity for 3TC-triphosphate, the mutant reverse transcriptase was shown to bind the 3TC-triphospate with a reduced affinity over the wild-type enzyme resulting in a 48.8 fold increase in resistance.
- Decrease the rate of incorporation of the NRTI-triphosphate over the natural nucleotide (eg. K65R, K70E, L74V and Q151M). In experiments with wild-type and mutant reverse transcriptase carrying the K65R mutation measuring the polymerisation activity for 3TC-triphosphate, the mutant reverse transcriptase was shown to reduce the rate of incorporation of 3TC-triphospate resulting in a 13.1 fold increase in resistance.
2. Excision pathway
These resistance mutations are amino acid changes in the primary structure of reverse transcriptase that facilitate the removal of the chain-terminators NRTI-triphosphate from the 3’ end of the DNA chain after it has been incorporated (eg. M41L, D67N, K70R, L210W, T215F/Y and K219Q).
The excision pathway is dependent on adenosine triphosphate (ATP) or pyrophosphate therefore mutations that increase the affinity of reverse transcriptase for ATP or increase the rate of removal of the analogue complex are favoured. Also changes in the translocation ability of the residues from the active site (N-site) to the post-translocation site (P-site) as well as the rate of dissociation of the template/primer from the enzyme can enhance the excision pathway.
Mode of Action – NNRTI
Non-nucleoside Reverse Transcriptase Inhibitors (NNRTIs) interfere with reverse transcription by directly binding to the enzyme and retarding its function. These are usually small chemical molecules with a long half-life.
This class is typically combined with two NRTIs to form first line HAART treatment in South Africa.
Mechanism of Action
Mechanisms of HIV antiretroviral drug action and drug resistance
Non-nucleoside reverse transcriptase inhibitors (NNRTI)
Mechanism of action
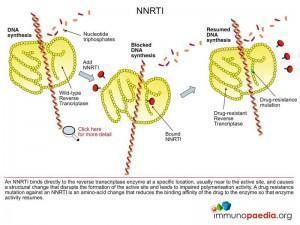
The NNRTI class of antiretroviral drugs are small hydrophobic chemical compounds that have high affinity for a hydrophobic binding pocket located near the active site of HIV reverse transcriptase. Binding of the drug to the enzyme results in a change in structural conformation of important residues required for optimal ability of the enzyme to catalyse DNA polymerisation.
Mechanism of Resistance
Mechanisms of HIV antiretroviral drug action and drug resistance
Non-nucleoside reverse transcriptase inhibitors (NNRTI)
Mechanism of resistance
These resistance mutations affect the binding of the drug to reverse transcriptase by changing the association and dissociation constants so that reverse transcription can be accomplished in the presence of the drug (eg. K103N and Y181C).
Mode of Action – Protease Inhibitors
Protease Inhibitors (PIs) are substrate analogues for the HIV aspartyl protease enzyme, which is involved in the processing of viral proteins. Once bound to the enzyme active site the enzyme is blocked from further activity. This inhibits the viral maturation process resulting in lack of functional virion formation.
These drugs are synergistic with reverse transcriptase inhibitors and are typically used in second line HAART treatment in South Africa.
Mechanism of Action
Mechanisms of HIV antiretroviral drug action and drug resistance
Protease inhibitors (PI)
Mechanism of action
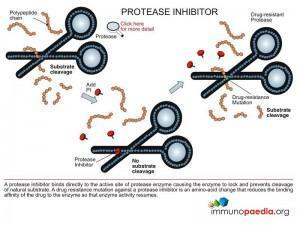
The protease inhibitors are small chemical compounds that mimic the natural peptide substrate of the enzyme, but do not allow for a cut to be made due to chemical modifications. Binding of the inhibitor to the active site of the enzyme prevents the enzyme from targeting its natural substrate since the inhibitor can only be released from the enzyme after the substrate has been cleaved.
Mechanism of Resistance
Mechanisms of HIV antiretroviral drug action and drug resistance
Protease inhibitors (PI)
Mechanism of resistance
These resistance mutations affect the binding of the drug to the active site of protease by changing the association and dissociation constants so that protease becomes available for proteolytic action in the presence of the drug.
Mode of Action – Integrase inhibitor
Integrase Inhibitors are a new class of drugs which target the HIV enzyme integrase. This is the enzyme responsible for the integration of viral genetic material into human DNA, a crucial step in the replication cycle of HIV.
Raltegravir (Isentress) is the first medicine to be approved in the class of antiretroviral drugs called integrase inhibitors. It was approved by the U.S. Food and Drug Administration (FDA) in October 2007. Raltegravir is approved for use in combination with other antiretroviral agents for the treatment of HIV-1 infection in treatment-experienced adult patients who have evidence of viral replication and HIV-1 strains resistant to multiple antiretroviral agents. It is not yet approved for people with drug-sensitive HIV strains, such as those starting antiretroviral therapy for the first time and has not been approved for use in children.
Mechanism of Action
Mechanisms of HIV antiretroviral drug action and drug resistance
Integrase inhibitors
Mechanism of action
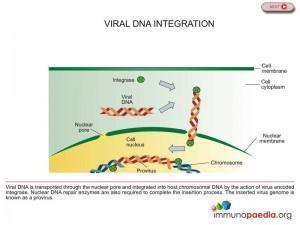
Integrase enzyme brings about the insertion of HIV DNA into human DNA, thereby helping to hide HIV’s DNA inside the host cell’s DNA (see figure 1). Once this provirus is formed the cell begins producing genetic material for new viruses. Raltegravir inhibits integrase from performing this essential function limiting the ability of the virus to replicate and infect new cells, thus preventing HIV DNA from meshing with healthy cell DNA (see figure 2). There are drugs in use that inhibit two other enzymes critical to the HIV replication process – protease and reverse transcriptase – but Raltegravir is the only drug approved that inhibits the integrase enzyme. For this reason Raltegravir is a significant milestone in the history of HIV/AIDS therapy.
Side Effects
The most commonly reported adverse experiences of any severity (mild, moderate or severe) regardless of drug relationship were diarrhea, nausea, headache and fever. Creatine kinase elevations were observed in subjects who received Raltegravir. Myopathy and rhabdomyolysis have been reported; however, the relationship of Raltegravir to these events is not known but should be used with caution in patients receiving concomitant medication known to cause these conditions.
Drug interactions
Based on the results of drug interaction studies and the clinical trials data, no dose adjustment of Raltegravir is required when co administered with other antiretroviral agents. Preclinical studies show that Raltegravir is not metabolized by cytochrome P450 enzymes. Caution should be used when co administering Raltegravir with strong inducers of uridine diphosphate glucuronosyltransferase (UGT) 1A1 (e.g., rifampicin) due to reduced plasma concentrations of Raltegravir.
Mechanism of Resistance
Mechanisms of HIV antiretroviral drug action and drug resistance
Integrase inhibitors
Mechanism of resistance
Genetic mutations in the viral integrase gene can produce a drug resistant integrase enzyme that can catalyse integration of the viral DNA.
Mode of Action – Entry Inhibitors
Entry Inhibitors interfere with the receptor-mediated entry of the virus into a cell. Two subclasses known as “fusion inhibitors” and “CCR5 antagonists”, are new classes of antiretroviral drugs used in combination therapy for the treatment of HIV infection.
This class of drugs interferes with the binding, fusion and entry process of HIV into a human cell.
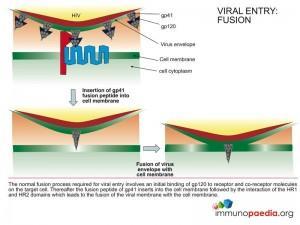 Mechanism of Action – Enfuvirtide
Mechanism of Action – Enfuvirtide
Mechanisms of HIV antiretroviral drug action and drug resistance
Entry inhibitors
(a) Subclass: fusion inhibitor (eg. Enfuvirtide)
Mechanism of action
The FDA approved fusion inhibitor Enfuvirtide is a peptide chain that mimics the structure of the HR2 region of gp41 that binds to the HR1 region and facilitates fusion of the viral envelope with the cell membrane. Binding of the inhibitor to the HR1 region prevents the HR2 region from access to HR1 and inhibits the fusion process.
 Mechanism of Resistance – Enfuvirtide
Mechanism of Resistance – Enfuvirtide
Mechanisms of HIV antiretroviral drug action and drug resistance
Entry inhibitors
(a) Subclass: fusion inhibitor (eg. Enfuvirtide)
Mechanism of resistance
These are resistance mutations in the HR1 region of gp41 that affect the binding affinity of the drug. A change in the association and dissociation constant of the drug allows the HR2 region to inititiate the fusion process.
Mechanism of Action – Maraviroc
Mechanisms of HIV antiretroviral drug action and drug resistance
Entry inhibitors
(b) Subclass: CCR5 antagonist (eg. Maraviroc)
Mechanism of action
The recently FDA approved CCR5 antagonist Maraviroc is a small chemical compound that binds to the external part of the CCR5 transmembrane receptor that serves as the co-receptor for virus entry. Binding of this inhibitor to CCR5 prevents HIV gp120 from access to the co-receptor and prevents the fusion process involving gp41 from proceeding.
Mechanism of Resistance – Maraviroc
Mechanisms of HIV antiretroviral drug action and drug resistance
Entry inhibitors
(b) Subclass: CCR5 antagonist (eg. Maraviroc)
Mechanism of resistance
- Amino acid substitutions and/or deletions in the co-receptor binding domain of gp120 (V3 loop) permits co-receptor binding to a different part of the CCR5 receptor which is unaffected by binding of the drug.
- Additional mechanisms of escape are genetic changes in the co-receptor binding domain of gp120 (V3 loop) that permits the use of an alternative co-receptor such as CXCR4 or other chemokine receptors or the positive selection of pre-existing CXCR4-using viruses