Recent advances in single-cell technologies and immune profiling have revealed just how diverse T-cell populations are. They exist far beyond the classic “CD4 helper” vs “CD8 cytotoxic” labels. But the way we name and classify these cells hasn’t kept up, leading to confusion, inconsistent definitions, and unnecessary complexity in the literature.
To address this, a group of immunology leaders have published a Consensus Statement setting out guidelines for how T cells should be named, defined and described in research (Figure 1). The paper argues for three core goals:
- Transparency in definitions. Every study should clearly state in the Methods section how they defined any T-cell subset (markers used, functional assays, gating strategy).
- Standardising existing subset terms. The statement offers “common experimental criteria” for popular T-cell subset names — helping ensure everyone uses the same definitions.
- Introducing a ‘modular nomenclature’ paradigm. Instead of forcing T cells into a handful of neat categories (e.g., “effector memory”, “exhausted”), the modular system describes cells by a set of biological properties (e.g., “antigen-experienced, high PD-1 expression, proliferative”, etc) in short descriptors.
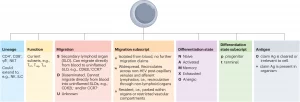
Figure 1: Modular T cell nomenclature v1.0. The figure provides a summary of the proposed modular nomenclature for T cells. Each column lists the optional descriptors for each parameter included in the modular nomenclature strategy v1.0. Ag, antigen; HEV, high endothelial venule; ILC, innate lymphoid cell; NK cell, natural killer cell; NKT cell, natural killer T cell.
Together, these guidelines aim to make T-cell research more transparent, more comparable across studies, and easier for researchers, students and clinicians to navigate.
- As T-cell biology becomes central to therapies, from vaccines to checkpoint inhibitors to CAR-T cells, consistent language is critical for translation across labs and clinics.
- With single-cell RNA and protein technologies, researchers are identifying dozens of previously hidden T-cell states. Without clear nomenclature, these proliferating definitions become confusing rather than helpful.
- The modular approach allows flexibility: rather than inventing a new subset name for every newly discovered state, researchers can describe cells via their functional features (modules) and only attach a formal “subset label” when widely agreed.
When reading or writing research on T cells:
- Look for the Methods section: how did the authors define “memory T-cell”, “exhausted T-cell”, etc?
- Expect to see descriptors such as “antigen-experienced CD8+ PD-1high KLRG1+” rather than just “exhausted T cell”.
- For clinicians and translational scientists: this should improve clarity when interpreting immune monitoring data, particularly in immunotherapy trials.
T cells are more varied than the old subset names suggest. This consensus statement offers a road map to standardise how we name and describe them, moving from rigid labels to a flexible, biology-based language that better reflects their true diversity and supports clearer communication across research and medicine.
Journal article: Masopust, D., et al. 2025. Guidelines for T cell nomenclature. Nature Reviews Immunology.
Summary by Stefan Botha










