In a recent study published in Science, Melani, et al., have reported on a several families of proteins in the body that may be used as a tool to predict which patients may be susceptible to rejection of a new organ transplant. This has the potential to aid clinicians in making informed decisions with regards to treatments.
The researchers were able to map specific indicators of immune cell proteins that underwent changes during transplantation within liver transplant recipients. The Blood Proteoform Atlas (BPA), outlines more than 56,000 exact protein molecules (called proteoforms) as they appear in 21 different cell types (Figure 1).
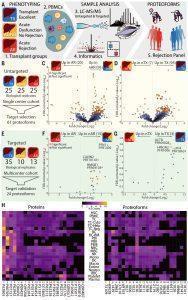
Figure 1: Quantitative top-down proteomics analysis of PBMC proteoforms from liver transplant recipients.(A) Workflow used to compare patients with transplant excellent (TX); with acute dysfunction, no rejection (ADNR); and with acute rejection (AR). (B) The number of patients whose PBMCs were analyzed in an untargeted fashion. Volcano plot showing differentially expressed proteoforms in (C) AR patients relative to non-AR (TX+ADNR) and (D) TX patients relative to non-TX (ADNR+AR). (E) The number of patients whose PBMCs were analyzed for targeted proteoforms. Volcano plot with the relative levels of the 24 targeted proteoforms in (F) AR versus non-AR and (G) TX versus non-TX. (H) Distributions of the normalized spectral (N.S.) counts of proteins (left) and proteoforms (right) observed in BPA cell types (Melani, et al., 2022).
There exists several unique forms of processed proteins (proteoforms) within the human body, form which millions of different proteins exist as a result of genetic variation and other mechanisms. In order to understand these proteoforms it has become a major goal of the initiative known as the Human Proteoform Project — discoveries about disease, aging and new therapeutics will accelerate.
In this paper, a specific focus was place on liver transplant patients. For monitoring symptoms of liver transplant rejection, physicians prescribe drug therapy in order to suppress the immune system and look for signs following treatment. Furthering our understanding of the mechanisms underlying rejection will eliminate the need for physicians to have to closely and strictly monitor these patients, whereby the chances of rejection is always a possibility, as we would be able to predict potential outcomes and modify our treatments accordingly to allow for better treatments and easier monitoring.
Using the BPA as a reference, the researchers sampled blood from participants in previous biomarker collection studies. They investigated and characterised which proteoforms played a role in the activation of immune response to the transplant and compared to patients without rejection.
They further developed a panel of 24 proteoforms from a previous study and looked at them in transplant recipient samples from across the country. They found a similar pattern.
This is a study with amazing potential as the use of a panel may help us identify patients who have no signs of rejection versus those who have very early evidence of rejection. This is will aid us in identifying rejection, several weeks before rejection actually happens allowing us to manipulate immunosuppression with more accuracy and under better guidance.
Journal article: Melani, R.D., et al., 2022. The Blood Proteoform Atlas: A reference map of proteoforms in human hematopoietic cells. Science.
Summary by Stefan Botha










