The study titled “Multilevel systems biology analysis identifies key immune response profiles and potential correlates of protection for the M72/AS01E vaccine against tuberculosis” applies an integrated computational framework to characterize host immune responses induced by the M72/AS01E tuberculosis vaccine. Using public RNA-seq data from a phase IIA clinical trial (GSE102574), the authors analyzed peripheral blood mononuclear cell (PBMC) gene-expression profiles from 18 vaccinated subjects collected at three timepoints: day 0 (baseline), day 31 (one day after second dose), and day 37 (one week after second dose).
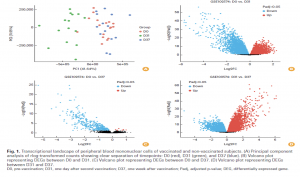
Differential-expression analysis revealed substantial transcriptional reprogramming, particularly at day 31, where 468 genes were differentially expressed compared to baseline. Gene-expression patterns shifted by day 37, indicating a transition from early innate responses to more refined adaptive signatures. Principal component analysis confirmed clear separation of samples by timepoint, reflecting robust vaccine-induced molecular changes.
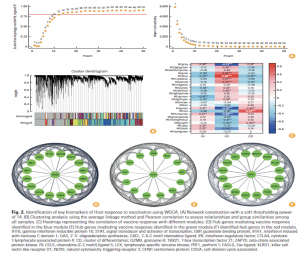
Using Weighted Gene Co-expression Network Analysis (WGCNA), the authors identified several co-expression modules correlated with vaccination. Among these, the blue, green, and red modules showed the strongest associations and were prioritized for downstream analysis. Network mining yielded 31 hub genes, many involved in interferon signaling, antiviral defense, and T-cell responses. Prominent hub genes included GBP1, GBP2, GBP4, GBP5, STAT1, STAT2, IFIH1, CXCL10, IRF1, IRF7, OAS1–3, PRF1, GZMB, TBX21, ZAP70, and others. These signatures reflect activation of type I interferon pathways, cytotoxic lymphocyte activity, and chemokine-mediated immune recruitment—hallmarks of effective vaccine responses.
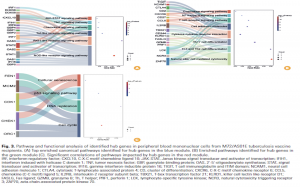
Functional enrichment analysis demonstrated significant involvement of RLR, TLR, and NLR signaling, JAK-STAT activation, Th1/Th2/Th17 differentiation, and T-cell receptor signaling, supporting the transition from innate to adaptive immunity after boosting.
The study incorporated machine-learning models (Random Forest, SVM, LDA, KNN) to classify timepoints and identify important predictive genes. Random Forest performed best, with high accuracy and biologically meaningful top-ranked features overlapping with WGCNA hub genes.
Finally, regulatory analysis integrating miRNA–TF–mRNA networks revealed multilayered control of key pathways, suggesting candidate regulators of vaccine-induced immunity.
Overall, the study provides a comprehensive systems-biology framework identifying immune modules, hub genes, and regulatory interactions associated with the M72/AS01E vaccine. These findings highlight potential correlates of protection that warrant further validation in larger and more diverse cohorts.
Journal article: Taofeek O.O., et al., 2025. Multilevel systems biology analysis identifies key immune response profiles and potential correlates of protection for M72/AS01E vaccine against tuberculosis. Clin Exp Vaccine Res.
Summary by Louis Ezediuno