In a recent paper, researchers have investigated several SARS-CoV-2 variants to describe how the innate immune system responds to infection (Figure 1). They specifically looked at the interferon response to SARS-CoV-2 infection.
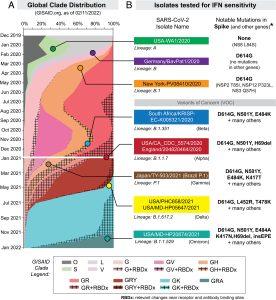
Figure 1: Selection of SARS-CoV-2 strains for IFN sensitivity studies. (A) Global distribution of SARS-CoV-2 clades. GISAID.org plotted the proportion of deposited sequences in designated clades against collection dates. The 10 isolates chosen are noted by colored dots. (B) SARS-CoV-2 strains selected for this study included representatives of lineages A, B, B.1, B.1.351, and B.1.1.7 (SI Appendix, Table S1). Lineage P.1 (which branched off from lineage B.1.1.28), B.1.617.2, and B.1.1.529 were added after the initial preprint submission and were evaluated for IFNβ and IFNλ1 sensitivity. Lineage B isolates encode the D614G mutation associated with increased transmissibility. *Amino acid mutations were relative to the reference hCOV-19/Wuhan/WIV04/2019 sequence (Guo, et al., 2022).
Much of the current research has been focused on neutralizing antibodies and the later stage of infection i.e., the adaptive immune response. Studies have shown that the virus has a unique way of adapting to evade innate responses as well.
The focus of this study, the group of proteins called interferons, are essential molecules in the innate immune system which may trigger a cascade of antiviral responses in cells quickly following infection. If primed properly, the innate immune system could reduce the viral load significantly upon initial infection.
In this paper they looked at 17 different human interferons and found that some interferons, more strongly inhibited SARS-CoV-2. It must be noted that they found that some later variants developed resistance to these innate mechanisms.
This study highlighted the importance of these molecules as a therapeutic target against SARS-CoV-2.
Journal article: Guo, K, et al., 2022. Interferon resistance of emerging SARS-CoV-2 variants. Proceedings of the National Academy of Sciences.
Summary by Stefan Botha










