A recent paper by Foix, et al., has reported on the potential of existing strains of SARS-CoV-2, and future variants to escape the immune system’s cytotoxic T cell response.
Genetically, HLAs determine the outcome of the T cell response in humans. This means that individuals share different and specific HLAs between them all in an effort to recognise pathogens based on their epitopes. Due to the vast amounts of differing HLA molecules, in addition to viral epitopes, to investigate the immune response of these specific alleles to each viral variant would be in immense task. Using a computational and systems biology approach to build prediction models may be a way of making this an easier and feasible task (Figure 1).
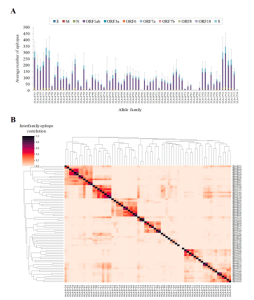
Figure 1: Number and degree of overlap between SARS-CoV-2 epitopes for different HLA-class I allelic families. (A) Average number of predicted HLA class I epitopes by allele family and protein. The standard deviation resulting from all proteins is indicated as a single error bar. (B) Hierarchical clustering and associated heatmap indicating the degree of inter-family epitope correlation. Color intensity expresses the Jaccard index for the epitope intersection between all family pairs. Perfect location match between epitopes calculated by netMHCIpan 4.1 EL with score 0.5 and rank 0.5 were utilized to calculate intersection and union. Intra-family conserved epitopes (50% alleles in the family by exact match) are in S1 Table (Fiox, et al., 2022).
In this present study, researchers first characterised an entire set of epitopes from the original strain of SARS-CoV-2 from Wuhan, China. Following this, the researchers analysed 118,000 different SARS-CoV-2 isolates from around the world to see for mutations in these epitopes. Nearly 50% of the epitopes were mutated in at least one existing isolate. They also found that the susceptible alleles were linked to geographic origin which was suggestive of a potential genetic pressure on the cytotoxic T cell response in these areas.
This approach has identified mutations within specific populations that warrant deeper interrogation and monitoring.
Journal article: Foix, A., et al. 2022. Predicted impact of the viral mutational landscape on the cytotoxic response against SARS-CoV-2. PLoS Computational Biology.
Summary by Stefan Botha










