In a recent discovery that may aid researchers in identifying new targets for cancer treatment, Belyaeva, et al., 2021 have discovered how immune cells protect themselves when they enter and squeeze between tissue cells.
The timing of when an immune cell invades a tumour cell is difficult to establish. In this present paper, the researchers turned to fruit flies to understand this question. During embryonic development of the fruit fly, macrophages, the dominant immune cell type, travel by means of tissue invasion. The process and time at which this is done has been studied by means of tagging macrophages with a fluorescent protein.
The authors shed light on the process by which cellular changes are needed for tissue invasion and which genes trigger these changes (Figure 1). They reported the assembly of actin filaments to facilitate this process and protect the invading cell. Through a complex cascade involving a variety of proteins, actin filaments are made denser and more connected to each other, forming a stable and protective coating. This leads to the protection of the immune cell’s nucleus from mechanical pressure as it invades the tissue. In addition, they reported, in vivo, that without the actin coating, it becomes difficult for immune cells to infiltrate surrounding tissue.
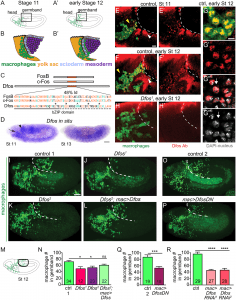
Figure 1.: The bZIP transcription factor Dfos acts in macrophages to facilitate their migration into the gb. Schematics of lateral (A) stage (St) 11 and (A’) early St 12 embryos. The boxed region magnified below indicates where macrophages (green) invade the gb after moving there from the head (B-B’). Macrophages sit on the yolk sac (yellow) next to the amnioserosa (black line) and then invade between the ectoderm (blue) and mesoderm (purple). (C) Dfos protein aligned with its human orthologs c-Fos and FosB; orange outlines the bZIP region that has 48% identity to both proteins: identical amino acids shown in orange, conserved ones in green. Stars indicate Leucines in the zipper; ^ the third leucine, which in Dfos is a methionine, a tolerated substitution [32]. The lower solid line indicates the basic domain and the dotted line the leucine zipper (ZIP). (D) In situ hybridization of St 11 and 13 embryos with a riboprobe for Dfos-RB (Fbcl0282531), which also detects all Dfos isoforms. Dfos RNA expression is enriched in macrophages (arrow) and the amnioserosa (arrowhead) before gb invasion but is gone thereafter. (E-H’) Confocal images of the boxed region in A from fixed embryos expressing GFP in macrophages (green) stained with a Dfos Ab (red). (E-F’, H-H’) A white dashed line indicates the gb edge. (E, F) The Dfos Ab stains (E) macrophages moving toward the gb at St 11 (yellow arrowheads), and (F) early St 12, as well as the amnioserosa (white arrowheads). (G) Higher magnification shows Dfos colocalizing with the nuclear marker DAPI (white). (H) No staining is detected in macrophages or the amnioserosa in the null Dfos1 mutant. (I-L) Lateral views of mid St 12 embryos from (I) the control, (J) the null allele Dfos1, (K) the hypomorphic allele Dfos2, and (L) Dfos2 with Dfos reexpressed in macrophages. (M) Schematic of St 12 embryo, gb region indicated by a black oval outline. (N) Quantitation reveals that both Dfos alleles display fewer macrophages in the gb. Reexpression of Dfos in macrophages in the Dfos2 hypomorph significantly rescues the defect. Control vs. Dfos1 p = 0.02 (30% reduction), Control vs. Dfos2 p = 0.017 (25% reduction), Control vs. Dfos2; mac>Dfos p = 0.334. (O-P) Lateral views of mid St 12 embryos from (O) the control, or (P) a line expressing a DN form of Dfos in macrophages. (Q) Quantification of macrophage numbers in the gb (see schematic) in the 2 genotypes visualized in O-P. p = 0.0002 (40% reduction). SD: 25, 25. (R) Quantification of macrophage numbers in the gb of the control and 2 different lines expressing RNAi constructs against Dfos in macrophages. Quantification of macrophage numbers in the gb for lines expressing one of 2 different UAS-Dfos RNAi constructs in macrophages. Control vs. mac>Dfos RNAi1 (TRiP HMS00254) or vs. mac>Dfos RNAi2 (TRiP JF02804), p < 0.0001 (54% or 52% reduction). SD: 32, 19, 29. The data in Q and R argue that Dfos is required within macrophages to promote gb tissue invasion. Embryos are positioned with anterior to left and dorsal up in all images, and histograms show mean + SEM throughout. Macrophages are labeled using srpHemo-Gal4 (“mac>”) driving UAS-GFP in E-H, UAS-GFP::nls in I-L and srpHemo-H2A::3xmCherry in O-R. ***p < 0.005, **p < 0.01, *p < 0.05. One-way ANOVA with Tukey post hoc was used for N and R, and unpaired t test for Q. The embryo number analyzed is indicated within the relevant column in the graphs. Scale bar: 50 μm in D, 5 μm in E-H, and 10 μm in I-L, O-P. The data underlying the graphs can be found in S1 Data. bZIP, basic leucine zipper domain; DN, dominant negative; gb, germband; RNAi, RNA interference; SD, standard deviation; SEM, standard error of the mean (Belyaeva, et al., 2022).
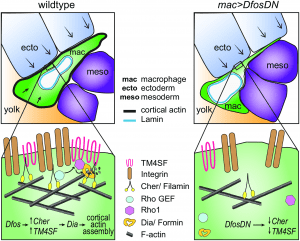
Figure 2: Model: Dfos increases actin assembly and cross-linking through the tetraspanin TM4SF and the filamin Cher to counter surrounding tissue resistance. We propose a speculative model for how Dfos tunes the cortical actin properties of Drosophila embryonic macrophages to aid their infiltration against the resistance of the surrounding gb tissue. We have shown that Dfos leads to an increase of the tetraspanin TM4SF and the filamin Cher. Filamins cross-link actin and have been shown to bind to RhoGEFs; Tetraspanins bind to Integrins, Rho GTPases, and Filamins in other systems (see S6 Fig). Thus, we hypothesize that in Drosophila macrophages, TM4SF and the filamin Cher could form a network at the cell surface of Integrin, actin, and upstream signaling molecules, recruiting Rho GEFs and leading to the activation of Rho1 GTPase and the actin polymerizing Formin Dia. Dia activation could occur through direct binding to active Rho1 and through direct interaction with TM4SF or Cher. Validation in Drosophila of all the protein interactions we propose awaits biochemical analysis. Through this pathway, a more cross-linked and dense F-actin network would form, aiding the macrophage in moving its cell body into the ecto–meso interface. The presence of Lamin around the nuclear membrane would not normally affect this process since the dense cross-linked cortical actin network would help macrophages withstand the load of the surrounding tissues. However, in the DfosDN-expressing macrophages, the loss of Cher and TM4SF would lead to reduced cross-linked actin levels at the cell cortex, making the stiffness of the nucleus the rate limiting step for macrophage infiltration of the gb tissue. Cher, Cheerio; ecto, ectoderm; gb, germband; mac, macrophage; meso, mesoderm (Belyaeva, et al., 2022).
“Our work also suggests a new perspective on the migration of some vertebrate immune cells. We find that altering lamin levels does not normally affect Drosophila macrophage tissue invasion…
A robust cell surface actin layer could allow long-lived cells or those not easily replenished to protect their genome as they move through resistant yet deformable environments. Embryonic Drosophila and vertebrate tissue-resident macrophages migrate into tissues during development, survive into the adult, and serve as founders of proliferative hematopoetic niches. Tissue-resident memory T cells migrate in response to infection in mature animals, are long lived, and are not easily renewed from the blood. Thus, the importance of nuclear mechanics for migration in challenging in vivo environments should be explored for a broader range of immune cells as well as the utilization of cortical actin as a strategy for genomic protection.”
Journal article: Belyaeva, V, et al., 2022. Fos regulates macrophage infiltration against surrounding tissue resistance by a cortical actin-based mechanism in Drosophila. PLOS Biology.
Summary by Stefan Botha










