In a recent study, researchers discovered that fighting diseases that take root inside our cells, like TB, requires the body’s process of removing old and injured cell components (Figure 1). If this natural process can be used in combination with novel therapies, it may offer an option to or enhance the use of antibiotics, particularly in situations where microbes have developed drug resistance.
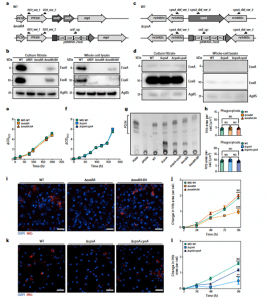
Figure 1: Characterization of Mtb ΔesxBA and Mtb ΔcpsA. a,c, Mtb esxBA- rv3874-75 locus (a) and cpsA-rv3484 locus (c) in Mtb WT and the respective deletion strains. Black half-arrows depict the primer positions (CmR, chloramphenicol resistance; ZeoR, zeocin resistance; Prom., groEL promoter). b,d, Western blot of EsxA and EsxB from total cell lysates and culture filtrates from Mtb WT, ΔRD1, ΔesxBA, ΔesxBA:BA (n = 3) (b) or Mtb WT, ΔcpsA and ΔcpsA:cpsA strains (n = 1) (d). Ag85 was used as a loading control. e, Growth curves of Mtb WT, ΔesxBA and ΔesxBA:BA. f, Growth curves of Mtb WT, ΔcpsA and ΔcpsA:cpsA strains. g, Thin-layer chromatography analysis of PDIM from Mtb WT, ΔcpsA, ΔcpsA:cpsA, ΔesxBA and ΔesxBA:BA cultures (n = 1). Purified PDIM and extracts from Mtb ΔPDIM were used as controls. h, Quantitative analysis of Mtb WT, ΔesxBA, ΔesxBA:BA (top) and Mtb WT, ΔcpsA, ΔcpsA:cpsA (bottom) area per single cell, 2 h post infection.Representative data of three independent experiments (n = 3 independent wells). Results are shown as mean ± standard error of the mean (s.e.m.). One-way ANOVA followed with Šídák’s multiple comparison test. NS, non-significant. i, Snapshot of live iPSDM infected with Mtb WT, ΔesxBA and ΔesxBA:BA 96 h post infection. Nuclear staining (blue) and Mtb-E2-Crimson (red). Scale bars, 50 μm. j, Quantitative analysis of Mtb replication after infection with Mtb WT, ΔesxBA and ΔesxBA:BA. Mtb area per cell was calculated as fold change, relative to 2 h post infection. Representative data of three independent experiments (n = 3 independent wells). Results are shown as mean ± s.e.m. One-way ANOVA followed with Šídák’s multiple comparison test. ***P < 0.001; NS, non-significant. k, Representative images of fixed iPSDM infected with Mtb WT, ΔcpsA and ΔcpsA:cpsA 96 h post infection. Nuclear staining (blue) and Mtb-E2-Crimson (red). Scale bars, 50 μm. l, Quantitative analysis of Mtb replication after infection with Mtb WT, ΔcpsA and ΔcpsA:cpsA. Mtb area per cell was calculated as fold change, relative to 2 h post infection. Representative data of three independent experiments (n = 3 independent wells). Results are shown as mean ± s.e.m. One-way ANOVA followed with Šídák’s multiple comparisons test **P < 0.002; NS, non-significant
In this research, they looked at genes important for bacteria’s ability to avoid autophagy, a process that cells use to self-destruct when under duress or contaminated. From induced pluripotent stem cells, which can develop into any form of cell in the body, they created human defence cells known as macrophages. They then altered the macrophage’s capacity for autophagy using genome engineering techniques. The bacterial infection took root when genes essential for autophagy were deleted from the cells and Mycobacterium tuberculosis was introduced, leading to increased bacterial replication within the engineered cells and widespread host cell death.
These findings support the idea that autophagy plays a significant part in preventing intracellular diseases like tuberculosis. If this pathway can be improved or reinforced, it may open new possibilities for combating antibiotic resistance by increasing the potency of currently available antibiotics or providing a treatment option in situations where microbes have developed resistance.
This novel process has created an opportunity to test drug substances that might be used to specifically increase autophagy. This won’t be a simple job, though.
Journal article: Beren Aylan, B., et al., 2023. ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages. Nature Microbiology.
Summary by Stefan Botha










