A comprehensive single-cell atlas reveals how tissue environment and time shape eosinophil identity across the body (Figure 1).
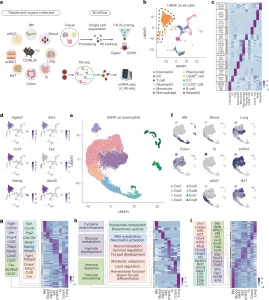
Figure 1: Single-cell transcriptomics analysis identifies tissue-specific eosinophil states. a, Schematic diagram of tissue processing, single-cell sequencing and antibody sequencing workflow. b, Integrated UMAP of all tissues with cell type annotations. DC, dendritic cell; ILC, innate lymphoid cell. c, Heatmap of DEGs across all cell populations. d, Expression of eosinophil marker genes. e, UMAP of eosinophil transcriptomes obtained from the BM, blood, lungs, colon, SI, scWAT, eWAT and BAT of C57BL/6 mice. f, Distribution of tissue origins projected on UMAP. g, Heatmap displaying tissue-specific DEG patterns. h, Tissue-originated pathway enrichment analysis shown by heatmap. i, TF regulatory network heatmap from SCENIC analysis across tissues (showing TF activation).
Eosinophils are best known for their roles in allergy and asthma, but these immune cells are far more versatile than once thought. They contribute to immune regulation, metabolism, tissue repair, and host defence. Despite their importance, a fundamental question has remained unanswered: how do eosinophils differ across tissues, and what governs their specialisation?
A new study provides the most detailed answer yet. Using cutting-edge single-cell technologies, researchers have created a transcriptomic and proteomic atlas of eosinophils across multiple mouse tissues, uncovering how local tissue signals and time spent in a tissue jointly determine eosinophil identity.
The research team combined:
- Single-cell RNA sequencing (to measure gene expression),
- High-dimensional surface proteomics (to profile cell-surface markers), and
- In vivo fate mapping (to track eosinophil lifespan and movement),
to analyse eosinophils from immune organs, barrier tissues, and metabolic sites.
This integrated approach allowed the researchers to follow eosinophils from their origins in the bone marrow through different stages of maturation and tissue residency.
One of the important findings was that eosinophil heterogeneity depends strongly on how long cells reside in a given tissue.
- In the small intestine, eosinophils are long-lived and diversify into multiple transcriptionally and phenotypically distinct subsets.
- In contrast, eosinophils in the lungs (short-lived) and colon (intermediate-lived) remain relatively uniform, showing far less specialization.
These results suggest that eosinophils continue to mature after entering tissues, with prolonged exposure to local cues driving diversification.
The study also identified trajectory-associated surface markers that mark eosinophil progression from bone marrow progenitors to long-term tissue-resident cells. These markers provide a practical toolkit for distinguishing eosinophil states in future studies, without relying solely on transcriptional profiling.
Together, the data support a unified model in which:
- Tissue-specific signals provide contextual instruction, and
- Duration of residency determines the depth of eosinophil maturation and specialization.
Eosinophils are implicated in a wide range of conditions, including asthma, inflammatory bowel disease, metabolic disorders, fibrosis, and cancer. Understanding which eosinophil subsets are present, and how they arise, is critical for interpreting their roles in both health and disease.
This atlas offers a molecular framework for resolving eosinophil diversity and sets the stage for:
- More precise disease models,
- Better interpretation of eosinophil-targeted therapies, and
- Identification of tissue- and state-specific therapeutic targets.
Eosinophils are not a single uniform cell type. Their identity is shaped by where they live and how long they stay there. By building a detailed single-cell atlas, researchers show that tissue residency time is a key driver of eosinophil maturation and diversity, providing new tools to understand their roles across health and disease.
Journal article: Hu., Y., et al. 2026. Temporal and spatial atlas of eosinophil specialization across tissues. Nature Immunology.
Summary by Stefan Botha










