When our immune system launches an attack, precision is everything. Natural killer (NK) cells and T cells act as the body’s elite assassins, releasing microscopic “toxic granules” that destroy virus-infected or cancerous cells. But how do these immune cells know exactly when and where to strike? A new study has uncovered a surprising link between lipid metabolism and the immune system’s ability to deliver its lethal cargo (Figure 1).
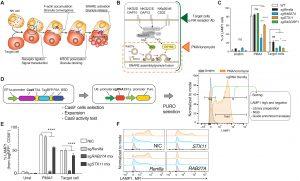
Figure 1: A genome-wide CRISPR KO screen to identify regulators of NK cell CG exocytosis. (A) Schematic of NK cell target cell recognition, lytic granule polarization, and killing function. (B) Differences in NK cell signaling events upon NK receptor ligation (K562 recognition/anti-NK receptor Ab, green) or chemical stimulation using PMA/I (orange). (C) Degranulation with indicated NK-92 cell lines comparing surface up-regulation of LAMP1 on unstimulated, PMA/I, and target stimulated cells. Bulk NK-92 cell lines were used for comparison. Data are shown as mean with SD. (D) Overview of FACS-based degranulation screen. Left: Generation of Cas9-expressing NK-92 cell line and consequent transduction with Brunello CRISPR library, selection, and expansion. Right: Exemplary FACS histogram showing gating strategy for LAMP1-stained PMA/I-activated NK-92 cells. LAMP1-negative (neg) and -high (hi) populations were sorted for further analysis of guide enrichment. Gating on the LAMP1+ (positive) population was used in all degranulation assays throughout this study to compare the efficiency. (E) Degranulation assay for NK92 Cas9-BFP cells transduced with lentiGuide. Puro vectors encoding control guides for Renilla, RAB27A (mix of three guides), and STX11 (mix of three guides). (F) Representative histogram stacks comparing surface LAMP1 up-regulation by noninfected cells (NICs) and sgRenilla, sgRAB27A, and sgSTX11 NK-92 cells quantified in (E). Data are shown as means ± SD and representative of at least two independent experiments. P values were calculated by two-way ANOVA with Bonferroni correction. Unstimulated (w/o, gray); stimulated with PMA/I (orange) or target cells (blue). *P < 0.05, ***P < 0.001, and ****P < 0.0001. ns, not significant.
Our immune system’s killer cells rely on the release of tiny, toxic granules that punch holes in infected or malignant cells. While scientists have identified several molecules critical for this process through the study of rare immune disorders, many details of this cellular choreography remained a mystery.
Using the team identified an unexpected set of genes essential for the precise release of these granules. To their surprise, many of these genes were tied to lipid metabolism, the same biological pathways that regulate how cells process and modify fats.
The study revealed that specific lipids help guide key proteins to the right place inside immune cells, ensuring the targeted release of cytotoxic granules. Without this lipid-based navigation system, immune cells lose their precision, reducing their ability to eliminate threats effectively.
Intriguingly, several of the identified molecules were previously known from neuronal biology, where they play roles in communication between nerve cells.
This study highlights the deep connections between metabolic and immune processes. By revealing how lipid metabolism influences the precision of immune cell attacks, the research bridges two seemingly distinct biological systems. Beyond advancing fundamental immunology, these insights hold clinical promise, improving the diagnosis of rare immune disorders and informing the development of next-generation cancer immunotherapies.
Journal article: Kalinichenko, A., et al. 2025. Protein palmitoylation and sphingolipid metabolism control regulated exocytosis in cytotoxic lymphocytes. Science Immunology.
Summary by Stefan Botha










