Regulatory T cells (Treg cells) are essential guardians of the immune system. They not only suppress harmful immune responses that can lead to autoimmune diseases, but they also play a key role in tissue repair by supporting wound healing and regeneration. Yet, to promote tissue recovery effectively, Treg cells must transform into tissue-resident Treg cells, a process that has remained poorly understood.
A new study described this transformation by uncovering distinct epigenetic changes that enable Treg cells to acquire tissue-healing functions (Figure 1).
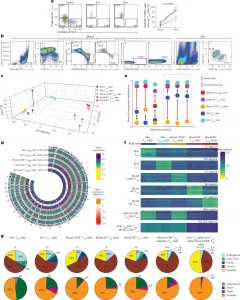
Figure 1: DNA methylation separates skin Treg and blood CCR8+ Treg cells from blood CD45RA+ Treg cells. a, Representative flow cytometry plot showing CD45RA−CCR8+ Treg cells among CD4+CD127−CD25+ Treg cells in human blood, fat and skin (left) and percentage of CD45RA−CCR8+ Treg cells among CD4+CD127−CD25+ Treg cells in human blood compared to skin (right; n = 6 healthy female donors; FACS Donors 1–6; median age: 46 years; range: 31–61 years). The P value was determined by two-tailed Wilcoxon signed-rank test. b, Sort layout showing isolation of blood CD45RA+ Treg, blood CD45RA+ Tconv, blood CCR8+ Treg, skin Tconv and skin Treg cells in one healthy female donor (FACS Donor 7; age unknown). Not all gates are shown. c, Principal component analysis of DNA methylation in blood CD45RA+ Treg cells (RA+ Treg cells), blood CD45RA+ Tconv cells (RA+ Tconv cells), blood CCR8+ Treg cells, skin Tconv cells and skin Treg cells isolated from blood and skin from nine healthy female donors (Tissue Donors 7, 10 and 11 and Blood Donors 3–8). d, Methylation level by genomic position and corresponding differences with respect to blood CD45RA+ Tconv cells in blood CD45RA+ Treg, blood CCR8+ Treg, skin Tconv and skin Treg cells in human donors as in c. Numbers in brackets indicate the average methylation level on autosomes and chromosome X. e, Schematic showing the extraction of cell-type signatures based on the largest numeric methylation gap between any two cell types (red arrows), which was required to be at least 0.15 and at least 1.5 times as large as the second-to-largest methylation gap (blue arrows) and resulted in the selection of signature regions; RA, CD45RA. f, DNA methylation in regions belonging to selected cell-type signatures as in e. Rows correspond to signature regions, and columns correspond to donors (n = 3 donors per cell type as in c). Numbers on the right indicate the number of regions per signature category. Signature nomenclature is based on the cell types that were hypomethylated in the corresponding signature regions. g, Distribution of cell-type signature regions from the signatures in e in genomic intervals defined by genes (top) and CpG islands (bottom). Numbers at the top indicate the number of regions per signature category. Data are representative of three or more independent experiments with three or more individual donors; TSS, transcription start site.
Treg cells are best known for maintaining immune balance, preventing overactive immune responses. But beyond immune suppression, they actively:
- Release tissue-healing molecules
- Support regenerative cells, including tissue stem cells
- Cooperate with both immune and non-immune cells to orchestrate wound healing
These diverse roles make Treg cells promising candidates for cell-based therapies aimed at restoring tissue function after chronic inflammation or severe injury.
However, to carry out these specialized tasks, Treg cells must undergo a functional reprogramming to become tissue-Treg cells. Until now, the precise molecular mechanisms driving this shift were unclear. Researchers focused on the epigenetic changes that accompany Treg cell differentiation.
Epigenetics refers to chemical modifications that regulate how genes are turned “on” or “off” without altering the underlying DNA sequence. One major mechanism is DNA methylation, where methyl groups are attached to specific DNA sites (CpG sites) to control gene activity.
By examining the DNA methylation patterns of Treg cells, the team identified a unique “epigenetic fingerprint” associated with tissue-Treg cells.
Key Discoveries
- Distinct DNA Methylation Signature
The researchers mapped the methylation profiles of 28 million CpG sites and found specific epigenetic patterns that uniquely define tissue-Treg cells. - Tracing Tissue-Treg Cells in the Blood
Surprisingly, this methylation “fingerprint” allowed the team to identify circulating precursors of tissue-Treg cells in the blood, revealing a potential way to monitor or harvest them for therapeutic use. - Involvement of Jumping Genes
They also discovered many changes in so-called “jumping genes” (transposable elements) – DNA sequences once thought to be inactive. These elements appear to play a role in regulating gene activity during Treg cell differentiation.
This research uncovers the molecular blueprint that guides Treg cells toward tissue-repair functions. Understanding this process opens the door to:
- Engineering Treg cells with enhanced wound-healing or anti-inflammatory capabilities
- Developing precision therapies for autoimmune diseases
- Promoting tissue regeneration after injury or chronic inflammation
Ultimately, this work provides a roadmap for harnessing tissue-Treg cells in regenerative medicine, turning these natural immune regulators into therapeutic tools for healing damaged tissues.
Journal article: Niklas Beumer., N., et al. 2025. DNA hypomethylation traits define human regulatory T cells in cutaneous tissue and identify their blood recirculating counterparts, Nature Immunology.
Summary by Stefan Botha










