Rheumatoid arthritis (RA) is a chronic autoinflammatory disease that is painful and can cause deformities. There are various environmental and genetic risk factors that have been associated with development of RA. One of the genetic risk alleles identified are involved in activation and differentiation of CD4 T cells. However, the precise CD4 T cell subsets involved in pathogenesis have not be identified. Identification of such subsets may lead to discovery of disease associated autoantigens.
Researchers aimed to identify RA disease-associated cell subsets using high dimensional single cell analysis. Previous studies have observed increased proportions of circulating CD28-CD4+ T cells in RA patients compared to controls. However, validating this result has been challenging, due to differences in clinical cohort, sample size and/or methodological variability among others. To identify other T cell immune phenotypes associated with this phenotype, researchers used a mass cytometry 22 parameter T cell panel to identify immune subsets enriched in RA patients compared to osteoarthritis.
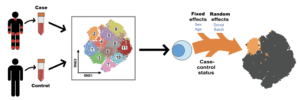
Fonseka et al., 2018. Figure 1: MASC overview. Single-cell transcriptomics or proteomics are used to assay samples from cases and controls, such as immunoprofiling of peripheral blood. The data are then clustered to define populations of similar cells. Mixed-effects logistic regression is used to predict individual cell membership in previously defined popula- tions. The addition of a case-control term to the regression model allows the user to identify populations for which case-control status is significantly associated.
Fonseka et al., used a robust statistical method called mixed-effects modelling of associations of single cells (MASC) to identify subsets associated with RA.
“MASC is a reverse association strategy where case-control status is an independent variable instead of a dependent variable.”
Using this method researchers were able to account for covariance due to technical and biological factors that could confound clustering of population. Additionally, clusters identified are not based on case-control status, but due to fixed (sex) and random (batch, donor etc.) effects. Using MASC and t-SNE analysis, they identified a population of CD27-HLA-DR+ CD4+ memory T cells that is enriched in RA patients, a population they were unable to identify using CITRUS (an alternative clustering platform). CD27-HLA-DR+ CD4+ T cells rapidly produced IFN-γ in response to non-specific stimulation, as well as express cytotoxic molecules granzyme A and perforin. Cytotoxic CD4+ T cells have been previously observed in RA synovial samples, thus this T cell phenotype could be a potential pathogenic T cell population.
In summary, Fonseka et al., identified a Th1-cytotoxic effector memory T cells population that is expanded in RA patients compared to controls using MASC and mass cytometry.
Journal Article: Fonseka et al.,2018. Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis. Science Translational Medicine.
Article by Cheleka AM Mpande












