TB caused by Mycobacterium tuberculosis (M.tb) is the leading cause of death worldwide by a single infectious agent. Approximately 10-20% people globally are exposed to TB, of which 5-10% of them will develop active TB disease. TB is diagnosed by either smear culture assays, chest x-ray and M.tb PCR tests such as GeneExpert, few tests are able to reliably diagnose subclinical disease, and there is currently no diagnostic that is available at the clinic/hospital.
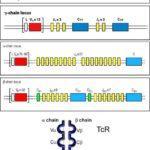
There are 4000 known M.tb antigens, current strategies exploring potential epitopes to target use IFN-Y ELISA to determine cellular responses to people infected (versus uninfected) to see if they elicit a good response to M.tb antigens (Bethel et al., 2008, JIMM). In addition, those individuals who control and clear infection or exhibit natural immunity offer insight into aspects of T cell recognition that may inform on better vaccine targets. A viable vaccine strategy to TB may involve inducing antigenic responses at different stages of the life cycle. Once potential TB antigens have been identified it is important to understand the T cell receptor (TCR) that will interact with the potential target. The specificity of the TCR to a specific antigen is determined by the germline encoded DNA sequence. This sequence is found within the VDJ region on the relevant chromosome coding for the alpha and beta chains. Recombination events enable TCR heterogeneity and enables recognition of a wealth of different peptides. Prof Thomas Scriba’s laboratory collected blood from M.tb infected individuals who progressed to TB, they then sorted T cells and conducted single cell sequencing of the TCR RNA (Musvosvi et al., unpublished).This enabled them to identify T cells that may play an important role in those who progress compared to those who don’t progress to TB disease.
Article by Esemu Livo Forgu, Holly Spencer and Kimone Fisher